A recent paper Social Epistasis Amplifies the Fitness Costs of Deleterious Mutations, Engendering Rapid Fitness Decline Among Modernized Populations provides a mathematical model to show the effects of behaviour of a subpopulation (carriers with behavioral mutations) on the total population. I was interested in playing with the parameters and so created a shiny app that would allow me to do so.
Assume co-mingling of a wild type population pop1 with a carrier population pop2. pop2 has deleterious behavioural mutation(s) that affect pop1 via social epistasis. Example behaviors that might have a negative impact on the population might be extreme altruism or free riding. The following equations from the paper describe the population growth of the two intermixed populations.
Population 1 (wild type) growth rate: $$pop1_{t+1} - pop1_t = (1 - p_{12})*birthrate1(pop1_t, pop2_t)*pop1_t - deathrate1*pop1_t$$ Population 2 (carrier) growth rate: $$pop2_{t+1} - pop2_t = birthrate2 * pop2_t - deathrate2 * pop2_t + p_{12} * birthrate1(pop1_t,pop2_t) * pop1_t$$ Birth rate of population 1: $$birthrate1(pop1_t, pop2_t) = birthsmax1* \frac{pop1_t}{pop1_t + pop2_t}$$To convert this to R code I utilize a vector indexed by the variable i which will represent the flow of time in generations. pop1[1] is the first generation, pop[2] the second etc. and more generally pop1[i] is followed by pop[i+1]. Likewise birthrate1, which is population size dependent, will be indexed by i, with a potentially unique value for each generation.
Start by defining variables and setting their values to the defaults used in the paper. pop 1 and 2 will be vectors
1 |
|
Loop over the generations, adjusting population sizes and birthrate1’s value, which is population size dependent.
1 |
|
Then plot
1 |
|
I would like to make this a Shiny app where I can dynamically change the variables and see the effect in the plot. Take the constants and wrap them into a reactive function. Here is server.R:
1 |
|
And the interface:
1 | shinyUI(fluidPage( |
Here is a reproduction of Figure 1 from the paper:
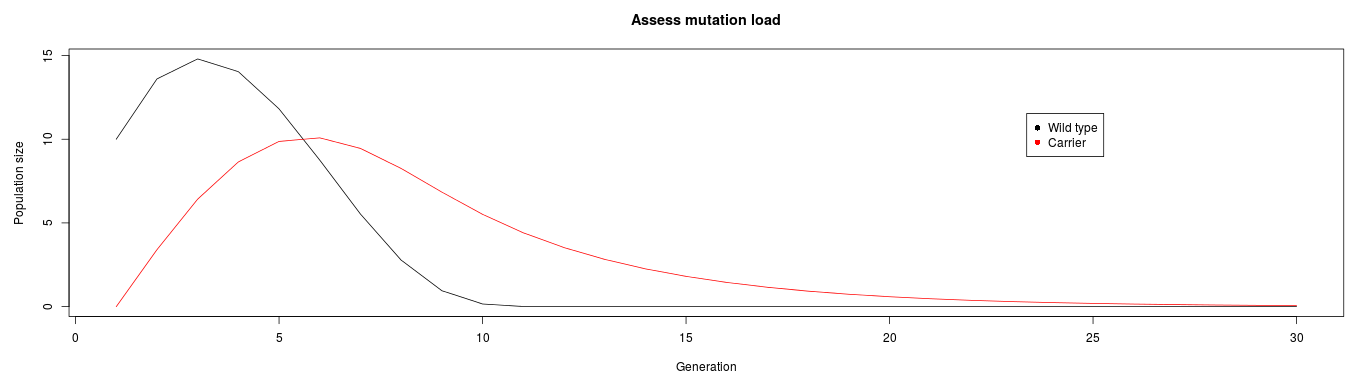
Here is the app on shinyapps.io. There is a monthly limit of free hours on shinyapps.io so you may have to come back if I have surpassed my limit.
Here is John Tooby with some commentary on purifying selection.
Ethnocentrism dominates humanitarianism.